What’s next for AlphaFold and the AI protein-folding revolution (Nature News)
ColabFold: Making protein folding accessible to all (Nature Methods)
Original AlphaFold paper
https://www.nature.com/articles/s41586-021-03819-2
- First computational method (neural network-based model AlphaFold) that can regularly predict protein structures from amino acid sequence alone, with atomic accuracy, even in cases in which no similar structure is known.
- Novel machine learning approach incorporates physical and biological knowledge about protein structure. AlphaFold network directly predicts the 3D coordinates of all heavy atoms for a given protein using the primary amino acid sequence and aligned sequences of homologues as inputs.
- AlphaFold is trained on protein chains in the PDB released before 2018-04-30.
- All-atom accuracy of AlphaFold in the CASP14 competition was 1.5 Å (1.4 Å is the width of a carbon atom).
The AlphaFold pipeline:
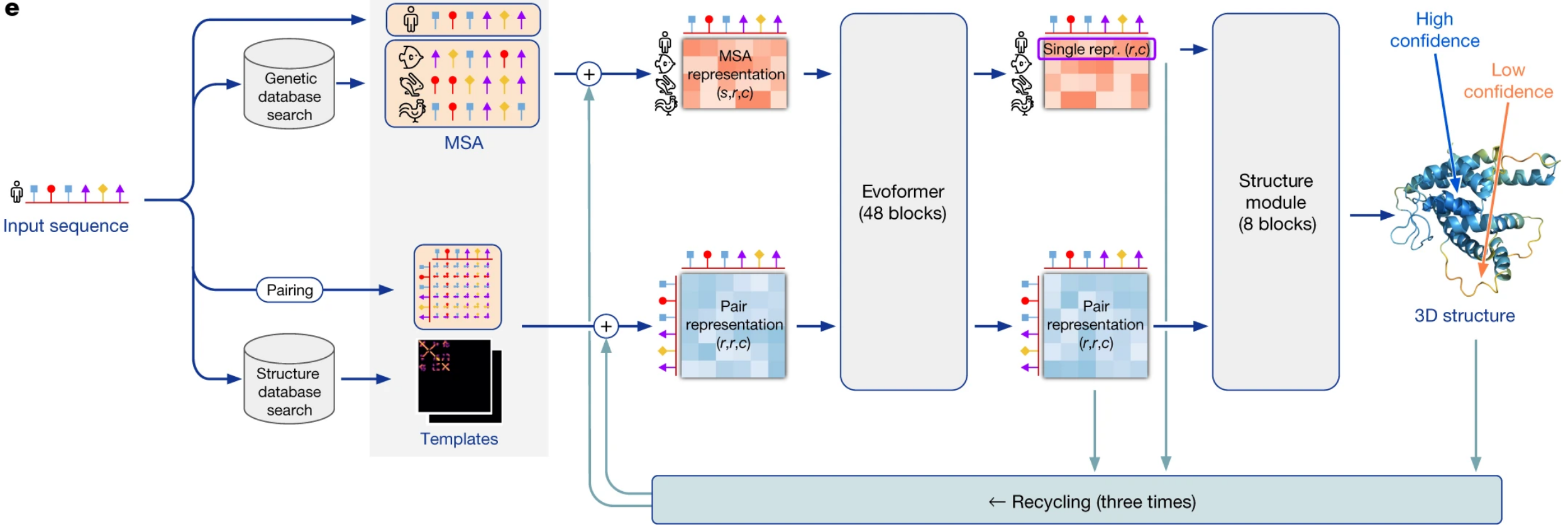
- Confidence measure = pLDDT(predicted local-distance difference test).
- Limitations: Model uses MSAs and the accuracy decreases substantially when the median alignment depth is less than around 30 sequences. AlphaFold is also much weaker for proteins that have few intra-chain or homotypic contacts compared to the number of heterotypic contacts; this occurs typically for bridging domains within larger complexes in which the shape of the protein is created almost entirely by interactions with other chains in the complex.
AlphaFold Protein Structure Database
- Search: search for queries based on protein name, gene name, UniProt accession or organism name. (Currently no BLAST/sequence-based searching supported, but you can use hte EBI Protein Similarity Search tool to seach AlphaFold DB based on a query sequence)
- Currently (as of July 2022) includes most of UniProt 2021_04 release, as well as proteomes of 20 model organisms and 48 complete proteomes. That means in total almost 215 Mio structures. Sequences that are not covered include ones that contain non-standard aas, are <16 aas or >2700 aas, and viral proteins. If your sequences aren’t included you can generate your own AlphaFold predicitions using DeepMind’s AlphaFold Colab notebook.
Example of structure page
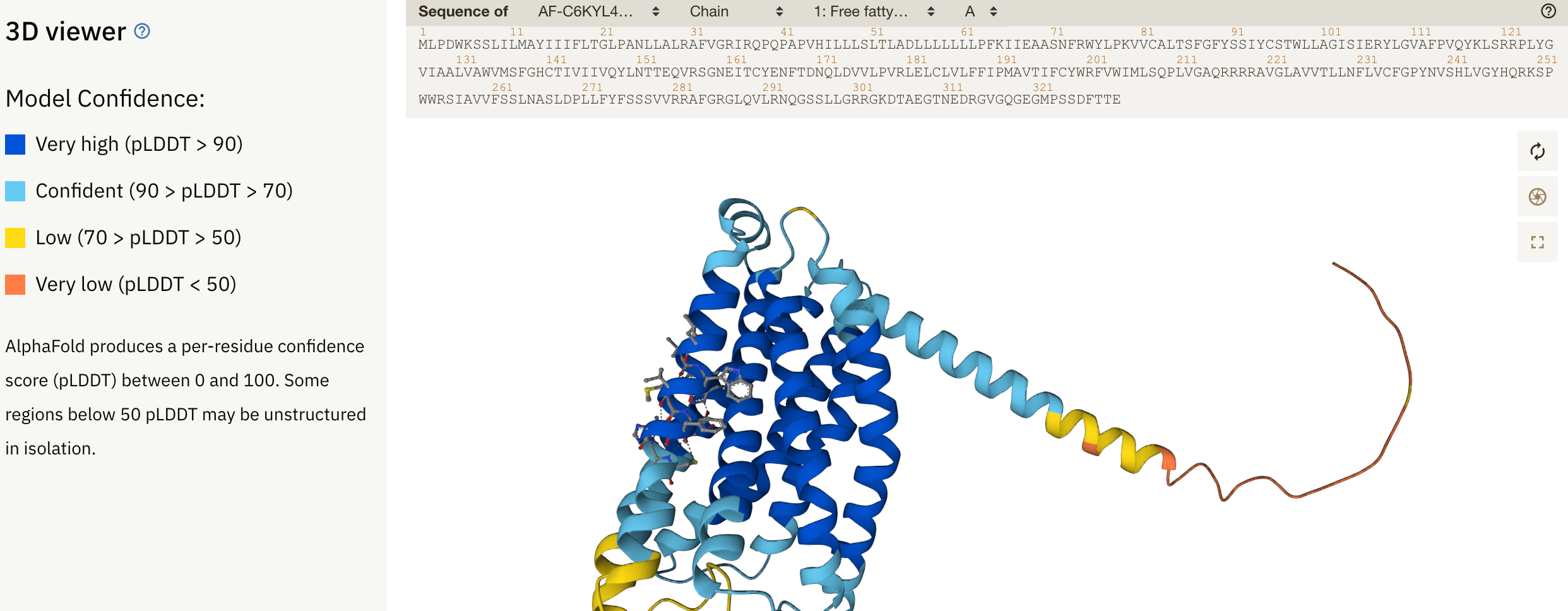
General structure and confidence
3D viewer: shows overall structure and confidence (pLDDT score; 100 = best, 0 = worst) in different parts of the structure. pLDDT > 90 are suitable for applications that require high accuracy, e.g. characterising binding sites. pLDDT < 70 should be treated with caution, and pLDDT < 50 should not be interpreted (shown as only a ribbon-like structure); these regions are often either unstructured in physiological conditions or only structured as part of a complex.
Click on aas in the sequence to zoom in and see side chains:
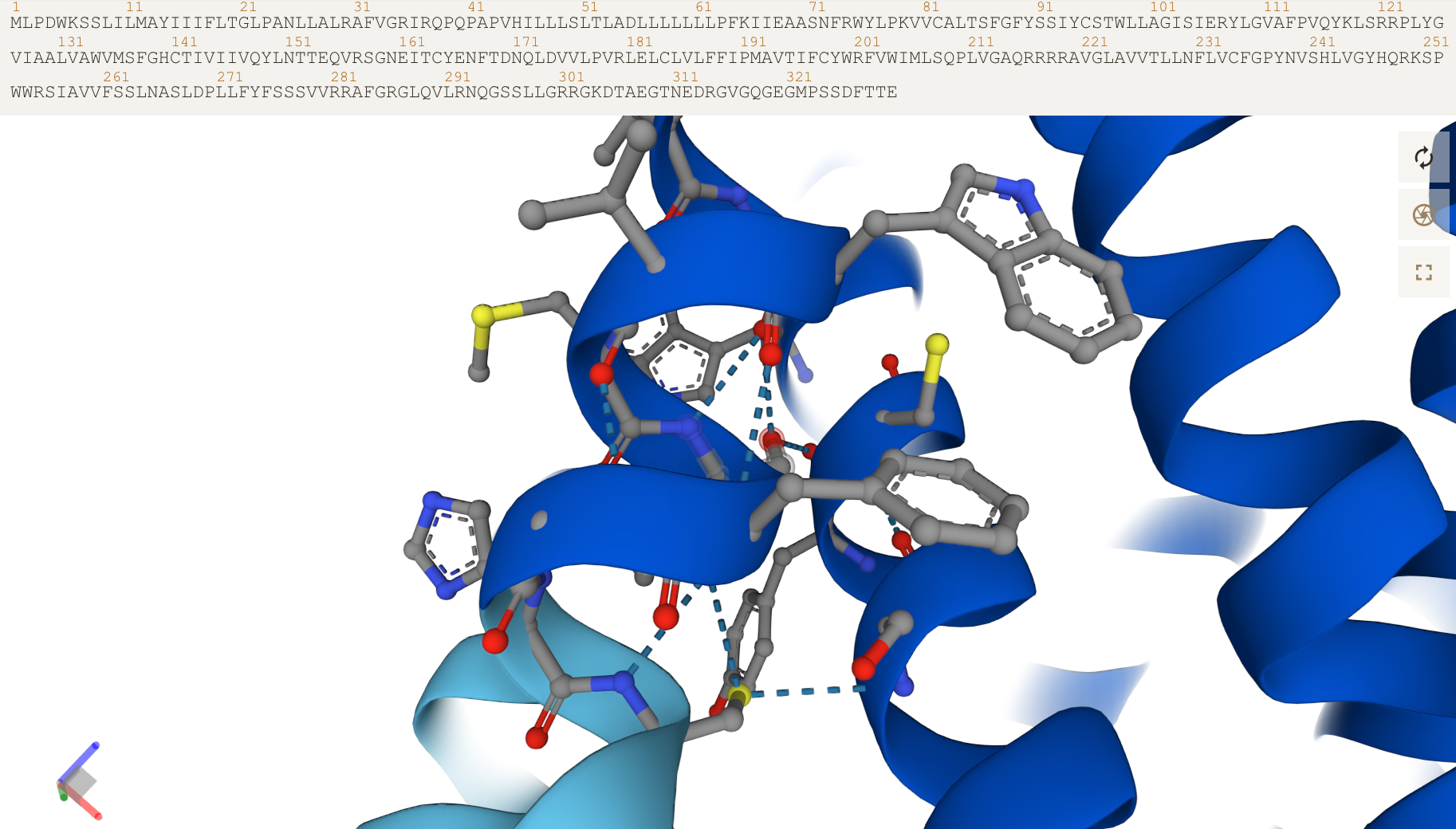
NB: PDB and mmCIF files that are downloadable contain coordinates for all regions, regardless of pLDDT score! It’s up to the user to interpret the model judiciously .
Relative position of domains
Predicted alignment error (PAE) is necessary to assess the confidence in the domain packing and large-scale topology of the protein:
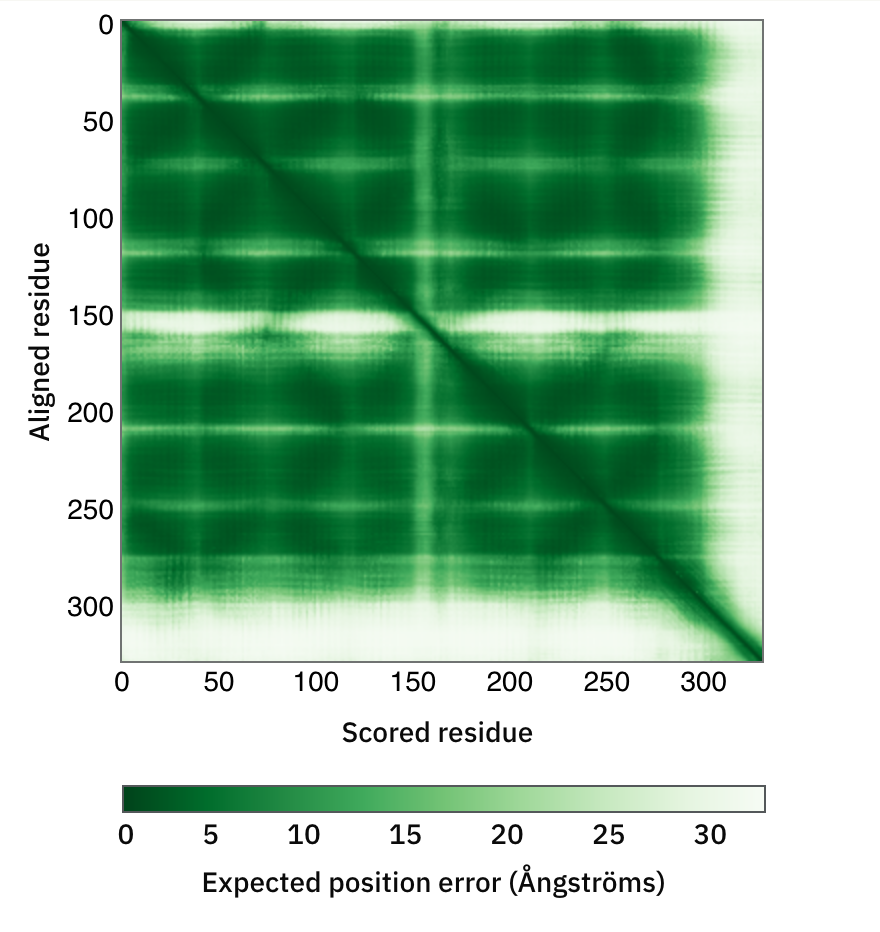
Colour at (x, y) indicates AlphaFold’s expected position error at residue x if the predicted and true structures were aligned on residue y. Low = good. If the PAE is high for residue pairs x, y from two different domains, then the relative positions and/or orientations of these domains in the 3D structure are uncertain and should not be interpreted.
PAE info can be downloaded as JSON file.
What AlphaFold does not support (yet)
- Intrinsically disordered or unstructured regions
- Effects of mutations. In particular, AlphaFold is not expected to produce an unfolded protein structure given a sequence containing a destabilising point mutation.
- Multiple conformations of a protein. AlphaFold usually only produces one of them.
- Predictions of positions of any non-protein components (e.g. cofactors, ligans, ions, post-translational modifications, DNA/RNA, metals, etc.). However, coordinates are often consistent with the expected structure in the presence of ions or cofactors.
Citing & Licence
If you use an AlphaFold DB prediction in your work, please cite the following papers:
Jumper, J et al. Highly accurate protein structure prediction with AlphaFold. Nature (2021).
All of the data provided is freely available for both academic and commercial use under Creative Commons Attribution 4.0 (CC-BY 4.0) licence terms.
AlphaFold Colab notebook
https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb
BFD: created by clustering 2.5 billion protein sequences from Uniprot/TrEMBL+Swissprot, Metaclust and Soil Reference Catalog Marine Eukaryotic Reference Catalog.